Now that you know about how blood works and the important role of hemoglobin, I’d like to explore the topic of sickle cell anemia as promised.
In the biology course Organisms and Environments we learned that the malaria virus is carried by a protist parasite called Plasmodium, which enters the human body via a mosquito vector. Malaria is the end product of a parasite chain, with mosquitoes carrying Plasmodium, which creates the disease symptoms.
 In Ecology and Population Biology, we learned that where malaria is present there is a higher prevalence of sickle cell anemia. Sickle cell anemia is almost non-existent in countries where malaria is not found, but something about the sickle cell disease produces resistance to malaria. In Genetics we learned that sickle cell anemia is a genetic disorder in which a change of one single nucleotide led to a novel amino acid substitution in the protein structure of hemoglobin. The disorder is autosomal recessive, so those that have two copies of the allele are fully affected, those that are heterozygous (two different copies) are only partially affected, and those that are homozygous dominant and don’t carry the mutated allele are not affected at all.
In Ecology and Population Biology, we learned that where malaria is present there is a higher prevalence of sickle cell anemia. Sickle cell anemia is almost non-existent in countries where malaria is not found, but something about the sickle cell disease produces resistance to malaria. In Genetics we learned that sickle cell anemia is a genetic disorder in which a change of one single nucleotide led to a novel amino acid substitution in the protein structure of hemoglobin. The disorder is autosomal recessive, so those that have two copies of the allele are fully affected, those that are heterozygous (two different copies) are only partially affected, and those that are homozygous dominant and don’t carry the mutated allele are not affected at all.Now, after all the build up between three of my biology courses, after years of asking “but how does sickle cell anemia produce resistance to malaria?” my question has finally been answered in Bio-Chemistry this year!
That single nucleotide change that is responsible for sickle cell anemia results in the inappropriate placement of a valine amino acid where a glutamate amino acid is supposed to be in the hemoglobin protein. Due to its polar qualities, the glutamate amino acid has no problem sitting on the outside of the hemoglobin protein facing the aqueous environment of the cell, and is therefore called “hydrophilic.” Valine, on the other hand, is a non-polar, uncharged amino acid and is said to be “hydrophobic.” Water tends to exclude this amino acid, pushing it away.
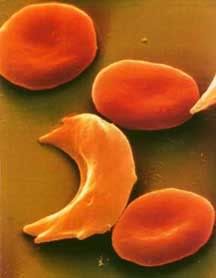 Hemoglobin in its relaxed state favors the binding of oxygen and has no hydrophobic sites revealed. When hemoglobin drops the oxygen and is in the tensed, deoxygenated state though, a hydrophobic niche is revealed, the perfect hiding place for that hydrophobic valine. If, by chance, the valine comes into contact with this site, it will not leave. Water excludes the hydrophobic molecules, forcing them to stick together. If enough hemoglobin valines find their way into this hydrophobic niche, aggregates of hemoglobin form in the erythrocyte (red blood cell), creating long hemoglobin strands. The cell will begin to form a sickle shape as a result of these strands and has potential to clot capillaries and even arteries if enough sickle cells are present. This is why it’s so dangerous for someone who has the sickle cell mutation to be in low oxygen environments; the less oxygen they are getting the higher the chance their hemoglobin will deoxygenate and aggregate to form sickle cells. This risk is so high in individuals who are homozygous for the mutation that they often die in childhood. People who are heterozygous for the mutation are somewhat better off. It’s still a risky disease, but their immune system does a pretty good job cleaning up those sickle shaped erythrocytes before they form clots.
Hemoglobin in its relaxed state favors the binding of oxygen and has no hydrophobic sites revealed. When hemoglobin drops the oxygen and is in the tensed, deoxygenated state though, a hydrophobic niche is revealed, the perfect hiding place for that hydrophobic valine. If, by chance, the valine comes into contact with this site, it will not leave. Water excludes the hydrophobic molecules, forcing them to stick together. If enough hemoglobin valines find their way into this hydrophobic niche, aggregates of hemoglobin form in the erythrocyte (red blood cell), creating long hemoglobin strands. The cell will begin to form a sickle shape as a result of these strands and has potential to clot capillaries and even arteries if enough sickle cells are present. This is why it’s so dangerous for someone who has the sickle cell mutation to be in low oxygen environments; the less oxygen they are getting the higher the chance their hemoglobin will deoxygenate and aggregate to form sickle cells. This risk is so high in individuals who are homozygous for the mutation that they often die in childhood. People who are heterozygous for the mutation are somewhat better off. It’s still a risky disease, but their immune system does a pretty good job cleaning up those sickle shaped erythrocytes before they form clots.Now, what does this have to do with malaria, you ask? Well, recall from the last article that there are a number of environmental signals that will cause hemoglobin to drop its oxygen. One of those signals is a low pH (acidity), as active tissues are often more acidic. When the parasite Plasmodium enters an erythrocyte the pH drops. The hemoglobin then drop their oxygen all at once, exposing the hydrophobic sites to the hydrophobic valines. They aggregate and cause the cell to sickle up, marking the cell for clean up by the immune system. The immune system is then able to sweep the parasite out, before it has a chance to reproduce!
Sources:
All info is from my coursework at UI
Images:
Sickle cells: http://www.defiers.com/scd.html
Plasmodium: wikipedia.com
 Oxygen wasn’t so popular at first, let me tell ya. The first organisms arose about 3.5 billion years ago when free oxygen was a rarity. They were anaerobic cells, and just as their name indicates, they had no use for oxygen. Originally, the process of glycolysis was used to break down organic compounds found in their environment. When the abundance of free organic compounds began to dwindle, photosynthetic organisms that could fix carbon dioxide (CO2) in the air to make their own organic compounds arose. A side effect of the ingenious design of photosynthesis just happened to be molecular oxygen (O2). There was very little O2 in the air at that time, except what they produced, and like us they tended to just let their waste build up (being unicellular, they didn’t exactly have the most creative waste management!). By about 2.5 billion years ago, the levels of oxygen began to poison these anaerobes. Dieing away in their own waste, these organisms seemed doomed…until a new organism hit the scene. One microbe’s trash is another microbe’s treasure, as they say, and an organism arose that took the idea behind photosynthesis, and practically put it in reverse. These organisms would utilize oxygen, and produce CO2 as a byproduct. Many of you have heard this story before, about how our ancestral mitochondrions saved the day and the entire earth from an overly oxidizing atmosphere. But the story goes deeper still.
Oxygen wasn’t so popular at first, let me tell ya. The first organisms arose about 3.5 billion years ago when free oxygen was a rarity. They were anaerobic cells, and just as their name indicates, they had no use for oxygen. Originally, the process of glycolysis was used to break down organic compounds found in their environment. When the abundance of free organic compounds began to dwindle, photosynthetic organisms that could fix carbon dioxide (CO2) in the air to make their own organic compounds arose. A side effect of the ingenious design of photosynthesis just happened to be molecular oxygen (O2). There was very little O2 in the air at that time, except what they produced, and like us they tended to just let their waste build up (being unicellular, they didn’t exactly have the most creative waste management!). By about 2.5 billion years ago, the levels of oxygen began to poison these anaerobes. Dieing away in their own waste, these organisms seemed doomed…until a new organism hit the scene. One microbe’s trash is another microbe’s treasure, as they say, and an organism arose that took the idea behind photosynthesis, and practically put it in reverse. These organisms would utilize oxygen, and produce CO2 as a byproduct. Many of you have heard this story before, about how our ancestral mitochondrions saved the day and the entire earth from an overly oxidizing atmosphere. But the story goes deeper still.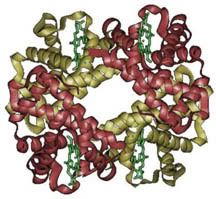 Hemoglobin is a protein with four subunits. It’s not all amino acids; each subunit also contains a prosthetic group, a non amino acid component that is essential to its function. This group, called the heme group, has a little something that oxygen loves very much: iron. Normally oxygen’s relationship with iron is, well, a little abusive. Oxygen takes electrons from iron atoms and leaves iron in an oxidized state, forming rust. The heme group takes precautions against such abuse. The iron is secured all around by a nitrogen ring that holds it steady with a distal histidine guarding its back. Histidine is one of the amino acids comprising hemoglobin and plays two necessary roles. For one, it hydrogen bonds the other end of the O2 molecule, keeping it in place. The other role is dealing with carbon monoxide. Carbon monoxide (CO) also has a love of iron, an affinity that is thousands of times greater than O2s. Luckily, the distal histidine gets in the way of what could be a beautiful relationship between the iron and CO. The distal histidine presses down into the plane where the CO would normally bind, forcing a bent conformation. CO is linear and unhappy in such a state, but O2, when bound to the iron, has no problem being bent and is comfortable at an angle.
Hemoglobin is a protein with four subunits. It’s not all amino acids; each subunit also contains a prosthetic group, a non amino acid component that is essential to its function. This group, called the heme group, has a little something that oxygen loves very much: iron. Normally oxygen’s relationship with iron is, well, a little abusive. Oxygen takes electrons from iron atoms and leaves iron in an oxidized state, forming rust. The heme group takes precautions against such abuse. The iron is secured all around by a nitrogen ring that holds it steady with a distal histidine guarding its back. Histidine is one of the amino acids comprising hemoglobin and plays two necessary roles. For one, it hydrogen bonds the other end of the O2 molecule, keeping it in place. The other role is dealing with carbon monoxide. Carbon monoxide (CO) also has a love of iron, an affinity that is thousands of times greater than O2s. Luckily, the distal histidine gets in the way of what could be a beautiful relationship between the iron and CO. The distal histidine presses down into the plane where the CO would normally bind, forcing a bent conformation. CO is linear and unhappy in such a state, but O2, when bound to the iron, has no problem being bent and is comfortable at an angle.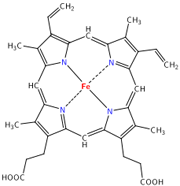 So the hemoglobin has packaged its oxygen. Very good, but it still must release this precious load to tissues that need it. How does it know when it’s at just the right place? Several things cue in the hemoglobin molecule. Active tissues are more acidic, with a higher concentration of CO2 and a lower concentration of O2. They also create the molecule 2,3-Bisphosphoglycerate (2,3 BPG) while breaking down glucose. Together, all of these molecules help to produce a conformational change in the hemoglobin protein complex. It releases it's oxygen and is now in the deoxygenated state and ready to pick up CO2. Check here to see the way hemoglobin moves to bind and release oxygen:
So the hemoglobin has packaged its oxygen. Very good, but it still must release this precious load to tissues that need it. How does it know when it’s at just the right place? Several things cue in the hemoglobin molecule. Active tissues are more acidic, with a higher concentration of CO2 and a lower concentration of O2. They also create the molecule 2,3-Bisphosphoglycerate (2,3 BPG) while breaking down glucose. Together, all of these molecules help to produce a conformational change in the hemoglobin protein complex. It releases it's oxygen and is now in the deoxygenated state and ready to pick up CO2. Check here to see the way hemoglobin moves to bind and release oxygen: 
 People have known about selection and evolution and used it to their advantage way before Darwin ever came on the scene. What in the world can I be speaking of? Why animal and plant breeding of course! Breeding is a fine example of evolution at work, but rather than being driven by natural selection it is driven by man’s selection. We breed animals with traits we find favorable until we have faster horses, better hunting hounds, juicier beef, or higher yield corn. Since it is well documented and we can even see the results of breeding within a lifetime, no one argues that this type of selection does not take place. But if we can do it, is it possible that nature can do the same thing if given the time?
People have known about selection and evolution and used it to their advantage way before Darwin ever came on the scene. What in the world can I be speaking of? Why animal and plant breeding of course! Breeding is a fine example of evolution at work, but rather than being driven by natural selection it is driven by man’s selection. We breed animals with traits we find favorable until we have faster horses, better hunting hounds, juicier beef, or higher yield corn. Since it is well documented and we can even see the results of breeding within a lifetime, no one argues that this type of selection does not take place. But if we can do it, is it possible that nature can do the same thing if given the time? In the natural world, organisms that have less favorable traits are less likely to survive and reproduce. Just as breeders may put down a slow horse to eliminate the ill suited genes from their stock, the slower gazelle may be put down naturally by hungry lions on the savannah. Aside from predator/prey type relationships, organisms may be less fit to survive a number of other natural challenges such as weather or other environmental conditions, disease, food shortages, competition, or they simply may not be able to find a mate willing to help them pass on their heritable traits. All of these are considered forms of natural selection.
In the natural world, organisms that have less favorable traits are less likely to survive and reproduce. Just as breeders may put down a slow horse to eliminate the ill suited genes from their stock, the slower gazelle may be put down naturally by hungry lions on the savannah. Aside from predator/prey type relationships, organisms may be less fit to survive a number of other natural challenges such as weather or other environmental conditions, disease, food shortages, competition, or they simply may not be able to find a mate willing to help them pass on their heritable traits. All of these are considered forms of natural selection. Cases like this happen all the time. Consider slow, dumb cattle being dragged down by wolves, crops being ravaged by disease, and let’s not forget the genetic abomination that is the chiwawa! It’s all a reminder of what man and nature can do to drive selection and create novel breeds or species.
Cases like this happen all the time. Consider slow, dumb cattle being dragged down by wolves, crops being ravaged by disease, and let’s not forget the genetic abomination that is the chiwawa! It’s all a reminder of what man and nature can do to drive selection and create novel breeds or species.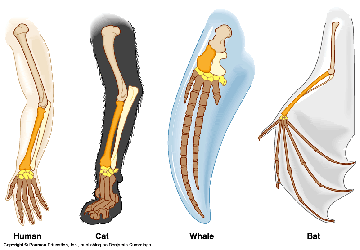

 Going deeper, we not only see that molecules like amino acids are active, but the very atoms and the bonds between them are as well. In the study of resonance structures in organic chemistry, we are shown that electrons are quite the flighty bunch. Take a look at this sulfate molecule below (also shown in 3D at left):
Going deeper, we not only see that molecules like amino acids are active, but the very atoms and the bonds between them are as well. In the study of resonance structures in organic chemistry, we are shown that electrons are quite the flighty bunch. Take a look at this sulfate molecule below (also shown in 3D at left):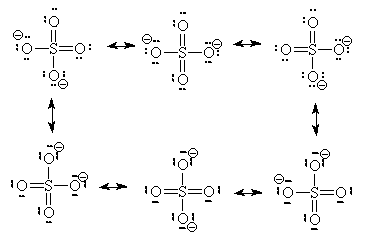



 Remember Gregor Mendel from high school biology? He was the monk with a certain love of peas who used probability to predict the expected ratio of traits in offspring. Mendel identified whether the physical traits of his pea plants were dominant (always expressed), partially dominant, or recessive (masked in the presence of dominant traits). If you were like me, you remember the "punnet square" method and getting stuck with any guy other than the one you liked, forced to make predictions about what eye color, brow line, etc., your offspring might have. Other uses of Mendel's findings are not always so traumatic. These methods can still sometimes be used to help predict the characteristics of offspring in breeding, research, and agriculture or anticipating a genetic disorder in humans. Although we now know that many traits are a result of multiple gene expression and most genes have more than just a dominant and a recessive allele (version), it was an amazing start and novel idea.
Remember Gregor Mendel from high school biology? He was the monk with a certain love of peas who used probability to predict the expected ratio of traits in offspring. Mendel identified whether the physical traits of his pea plants were dominant (always expressed), partially dominant, or recessive (masked in the presence of dominant traits). If you were like me, you remember the "punnet square" method and getting stuck with any guy other than the one you liked, forced to make predictions about what eye color, brow line, etc., your offspring might have. Other uses of Mendel's findings are not always so traumatic. These methods can still sometimes be used to help predict the characteristics of offspring in breeding, research, and agriculture or anticipating a genetic disorder in humans. Although we now know that many traits are a result of multiple gene expression and most genes have more than just a dominant and a recessive allele (version), it was an amazing start and novel idea. The famous Thomas Morgan (left) began his research career shortly after discovery of Garrod and others. Because humans don’t reproduce frequently and you certainly can't tell them who to breed with (unless they are high schoolers engaged in hypothetical crosses), Morgan thought his work would be better suited to a model organism:
The famous Thomas Morgan (left) began his research career shortly after discovery of Garrod and others. Because humans don’t reproduce frequently and you certainly can't tell them who to breed with (unless they are high schoolers engaged in hypothetical crosses), Morgan thought his work would be better suited to a model organism:  Later, in 1931, Harriet Creighton and Barbara McClintock (left) used corn to prove the physical existence of chromosomes and recombination. Using a chromosomal mutation in corn that made the ends of the chromosome distinguishable from one another under the microscope (one end with a “knob” and the other with extra length); they were able to show that recombination actually occurs. Genes more closely associated with the knobby end would recombine less frequently with one another and more frequently with those on the elongated end. McClintock’s research continued to contribute to genetics well into the 60’s, including her 1948 discovery of transposons (“mobile genes”) which she received a Nobel Prize for in 1983. Recombinant frequencies as well as other techniques are still used today to create relative genetic maps that aid in the sequencing process.
Later, in 1931, Harriet Creighton and Barbara McClintock (left) used corn to prove the physical existence of chromosomes and recombination. Using a chromosomal mutation in corn that made the ends of the chromosome distinguishable from one another under the microscope (one end with a “knob” and the other with extra length); they were able to show that recombination actually occurs. Genes more closely associated with the knobby end would recombine less frequently with one another and more frequently with those on the elongated end. McClintock’s research continued to contribute to genetics well into the 60’s, including her 1948 discovery of transposons (“mobile genes”) which she received a Nobel Prize for in 1983. Recombinant frequencies as well as other techniques are still used today to create relative genetic maps that aid in the sequencing process. It would take longer before technology would advance in such a way to prove the physical characteristics of genes and pave the road for sequencing of whole genomes. 1947 papers by Chargaff revealed that nucleotides adenine and thymine, guanine and cytosine always appeared in equal proportions suggesting possible pair bonding. In 1948 Linus Pauling (my bio-chem instructor’s favorite chemist) discovered the alpha helical shape of proteins which could potentially be applied to nucleotides. Later x-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin showed the possibility of DNA’s helical form. Putting all of this insight together, Francis Crick and James Watson (left) were able to publish the first structural model of DNA in 1953.
It would take longer before technology would advance in such a way to prove the physical characteristics of genes and pave the road for sequencing of whole genomes. 1947 papers by Chargaff revealed that nucleotides adenine and thymine, guanine and cytosine always appeared in equal proportions suggesting possible pair bonding. In 1948 Linus Pauling (my bio-chem instructor’s favorite chemist) discovered the alpha helical shape of proteins which could potentially be applied to nucleotides. Later x-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin showed the possibility of DNA’s helical form. Putting all of this insight together, Francis Crick and James Watson (left) were able to publish the first structural model of DNA in 1953.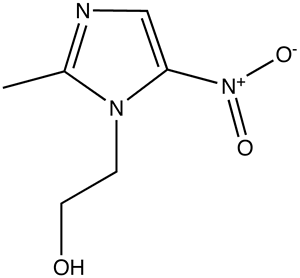
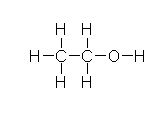 In the liver, alcohol (right) is broken down by alcohol dehydrogenases into acetaldehyde with the help of NAD by the following reaction: CH3CH2OH + NAD+ → CH3CHO + NADH + H+. Next, the acetaldehyde interacts with the acetaldehyde dehydrogenase (also in the liver) to form acetic acid (CH3-COOH, a very weak acid with many biological functions). Metronidazole is believed to obstruct the acetaldehyde dehydrogenase enzyme in much the same way disfulfiram, an anti-alcoholism drug, does. This allows excess acetaldehyde to build up in the body, 5-10 times as much acetaldehyde as one would normally experience after consuming an equal amount of alcohol with out the metronidazole. Since acetaldehyde is the primary cause of hangover, one can experience severe hangover-like symptoms including hot flash, shakes, nausea, and stomach cramps with even the slightest alcohol intake when combined with metronidazole.
In the liver, alcohol (right) is broken down by alcohol dehydrogenases into acetaldehyde with the help of NAD by the following reaction: CH3CH2OH + NAD+ → CH3CHO + NADH + H+. Next, the acetaldehyde interacts with the acetaldehyde dehydrogenase (also in the liver) to form acetic acid (CH3-COOH, a very weak acid with many biological functions). Metronidazole is believed to obstruct the acetaldehyde dehydrogenase enzyme in much the same way disfulfiram, an anti-alcoholism drug, does. This allows excess acetaldehyde to build up in the body, 5-10 times as much acetaldehyde as one would normally experience after consuming an equal amount of alcohol with out the metronidazole. Since acetaldehyde is the primary cause of hangover, one can experience severe hangover-like symptoms including hot flash, shakes, nausea, and stomach cramps with even the slightest alcohol intake when combined with metronidazole.