
What’s in a Name?
If you’ve seen my closet, you know I’m an organization enthusiast. All the hangers are color-coded, which is fairly easy when over half your cloths are black and a third are white, but I still have those precious few blue and red hangers, even a pink one for one salmon colored sweater and a green for my emerald blouse. The shirts follow a logical progression—starting with the warmest, longest sleeves and moving down to tank tops, the colors undergoing an easy shift from black to green to blue to plum to red to auburn to brown to grey then finally white. That’s not to say I’m a neat freak—my house is tidy but certainly not spotless—it’s just that if something is to be put in its place, I like a good logical order to it.
So it’s no surprise then that I’m a fan of nomenclature. Although I have my qualms with the Linnaean system for classifying organisms it does provide an arbitrary hierarchical order to things.
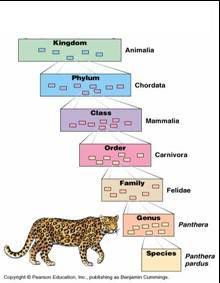 In the old days it was a simple regression from kingdom to phylum to class to order to family to genus to species. Now, thanks to new discoveries of organisms that just didn’t quite fit the bill, we begin with Domain, then move down to kingdom and so on. There are now sub and super everything (e.g.: superphylum and subphylum) to help further group organisms. Questions always arise of course as to who is more related to whom and where do we draw the line. My biggest qualm, and currently a topic of debate among nomenclature enthusiasts, is that the system is based primarily off of bone homology and other physical features, rather than possibly more sophisticated genomic methods. Genetic homology and analogy can tell a lot about the origins of an organism, and I feel it more accurately represents the relatedness of organisms than morphology.
In the old days it was a simple regression from kingdom to phylum to class to order to family to genus to species. Now, thanks to new discoveries of organisms that just didn’t quite fit the bill, we begin with Domain, then move down to kingdom and so on. There are now sub and super everything (e.g.: superphylum and subphylum) to help further group organisms. Questions always arise of course as to who is more related to whom and where do we draw the line. My biggest qualm, and currently a topic of debate among nomenclature enthusiasts, is that the system is based primarily off of bone homology and other physical features, rather than possibly more sophisticated genomic methods. Genetic homology and analogy can tell a lot about the origins of an organism, and I feel it more accurately represents the relatedness of organisms than morphology.Linnaeus aside, there are bigger problems lurking within the naming of genes themselves. Sonic hedgehog gene is a classic example. It may be cute, but it's not so cool when you're a doctor and you have to tell a pateint their baby died because of a "sonic hedgehog" mutation or if you have a debilitating disorder involving sonic hedgehog and you want people to take you seriously. Although quaint gene names like "sonic hedgehog," "space cadet,' and "cheap date" are funny, in addition to being slightly insulting to those who have serious genetic conditions they are also not very descriptive of the gene. They often describe one version of a mutation, or a knockout phenotype rather than what the gene actually codes for. For example, zebrafish larvae who have a knock-out mutation for space cadet display poor orienting behavior so that when you tap them they just swim around in a confused circle (which is really quite cute, check out these links for the wild type and mutant escape responses), but this does not say the function of the gene. Similarly, hedgehog gene knockouts produce a spiky appearance in flies but many mutations in mammals would suggest "Cyclops" as a more appropriate name based off of mammalian mutations. Most genes, though, are not nearly so cleverly named as sonic hedgehog or space cadet. Typically the gene gets a set of numbers and/or letters, which could mean anything from something biologically relevant to the initials of the person who discovered it. This is no good, a set of random numbers is far worse than naming based on even knock out mutant phenotypes. But how do we go about finding a better route?
I love IUPAC, the international system for naming molecules. There can be no mistake as to the structure of your molecule if it is a small metabolite and properly named. If I were to endow anyone with this project of designing a genetic nomenclature system, it would be them. I do have some ideas of my own though. I rather like the name of one or the genes I’m working with now—raldh2. It’s sensible and descriptive. Raldh2 codes for retinaldehyde dehydrogenase 2. Dehydrogenases are enzymes that do just as their names indicate (if you are familiar with organic chemistry)—they transfer protons and a pair of electrons to an acceptor. In this case the substrate is retinaldehyde and this enzyme is just one of 2 or more. As you can see, it’s easier to name a gene appropriately when we have a clearly defined protein product whose function is known. Until the protein product and something about their function is known, I think it is reasonable to temporarily name genes by their location on the chromosome of the organism they are being studied in (mouse, fly, human, or whatever it may be) until such a time as the functions are known. Once the protein product has been discovered, renaming genes like LOC643921 to something more akin to their actual function—such as say Yphos11 for tyrosine-protein phosphatase 11, would be ideal.
Now, back to my beef with sonic hedgehog gene. Sonic hedgehog would present a problem as the protein is named for the gene, so we cannot work off that. To further complicate things, the protein has many functions in an organism. If I were to name it though, I might try something like ZnMophLigand for zinc morphogen ligand, since it contains a zinc prosthetic group, acts as a ligand which binds to certain cell membrane proteins, and it is a morphogen (involved in regulating cell differentiation by the creation of diffusion gradients).
I know my knowledge of biochemistry is far from adequate for the creation of a good nomenclature system for genes. Still, I would like to see and would strongly support an effort to begin creation of a logical, international system of nomenclature for genes. No more crazy numbers, no more joshing around, just a clear, concise description of the gene’s biological relevance right in the name.
Sources:
http://www.internetadsales.com/modules/wfsection/index.php?category=7
http://tosaweb.ncsd.k12.wy.us/faculty/lbell/Taxonomy/Taxonomy.htm#slide0003.htm
Space cadet: http://dev.biologists.org/cgi/content/full/128/11/2131/DC1


3 Comments:
You think Genes are mess? Try Microbes once they switched to 16s ribosomal unit mapping and GC ratios as primary methods of classification. As it turns out, many of the organisms they had classed together, though similar in terms of enzyme production and nutritional needs, were completely different when it came to their actual genetic material. There are so many things in my lab manual that have now been completely changed, to the point where TA's will have to modify sample labels on the spot the day of class to make sure everything jives with current nomenclature.
By Anonymous, at 12:46 PM
Anonymous, at 12:46 PM
I think, more important than which system is used, is that everybody use the same system. For example, while Linnaean nomenclature is far from perfect, at least it's fairly consistent in its imperfection.
By Anonymous, at 3:53 PM
Anonymous, at 3:53 PM
Interesting ideas Kim. Your stuff is definently interesting to read...very refreshing perspective.
By Anonymous, at 4:43 PM
Anonymous, at 4:43 PM
Post a Comment
<< Home