As mentioned before, my primary work at the lab involves trying to gain insight into genes, their expression and their interactions. One challenge is telling when and where a gene is being expressed. A great indicator of gene expression that we commonly look for is the very first product of expression, mRNA. It is impossible to tell if a certain gene's mRNA is being produced just by looking under a light microscope, so we must utilize methods that make mRNA easier to visualize. A great way to do this is to make probes for the mRNA. Probes are complimentary RNA strands designed to hybridize to the mRNA of interest. Probes contain special nucleotides that are either fluorescently labeled, radioactively labeled, or, in our case, contain antigens for antibody labeling. This allows us to see where genes are being expressed easily under a microscope.
~Raising a Plasmid~
 Last week I received a letter in the mail containing a simple piece of filter paper wrapped in plastic. No, not another birthday card, this was my plasmid, the complimentary DNA I needed to make RNA probes. How we get that DNA from a piece of filter paper, create thousands of copies, and eventually make probe is an interesting technique that employs the services of microbiology’s favorite bacteria: Escherichia coli.
Last week I received a letter in the mail containing a simple piece of filter paper wrapped in plastic. No, not another birthday card, this was my plasmid, the complimentary DNA I needed to make RNA probes. How we get that DNA from a piece of filter paper, create thousands of copies, and eventually make probe is an interesting technique that employs the services of microbiology’s favorite bacteria: Escherichia coli. Don’t let the name scare you. Some strains of E. coli live peacefully in your intestine, while others, especially the dreaded 0157:H7 strain found on spinach leaves the other week, will make you horribly ill. In the lab, E. coli is the cattle of the microbiology world. We grow and raise them and they, in turn, produce product for us. Of course, we only use harmless, competent E. coli stains. Never, not even in all the years of practical jokes against unsuspecting undergrads, have we even touched the 0157:H7 strain.
Don’t let the name scare you. Some strains of E. coli live peacefully in your intestine, while others, especially the dreaded 0157:H7 strain found on spinach leaves the other week, will make you horribly ill. In the lab, E. coli is the cattle of the microbiology world. We grow and raise them and they, in turn, produce product for us. Of course, we only use harmless, competent E. coli stains. Never, not even in all the years of practical jokes against unsuspecting undergrads, have we even touched the 0157:H7 strain.The E. coli we use are said to be competent because they have the amazing ability to take-up DNA from their environments. The process of taking up DNA from the environment is called transformation, something competent cells are, well, competent at.
Once the plasmid has been eluted from the filter paper, it is introduced to the E. coli cells in a heat bath for half a minute. The heat makes the bacterial membranes more permeable, and it is hoped that many of the cells will take up the plasmid DNA in a marvelous transformation at this point. We then allow them to grow and reproduce for a while, undisturbed.
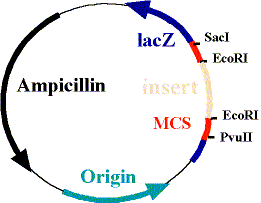
The plasmid is ingeniously designed. It contains the complimentary base pairing to our mRNA of interest, but it also contains an ampicillin resistance gene. This is essential, because it is the only way we are able to select for bacteria containing the plasmid. You can see a picture of a typical plasmid below, the "insert" being where our probe DNA would go and the ampicillin section represents the ampicillin resistance gene:
After letting them grow for a day, we plate the bacteria on medium containing the antibiotic ampicillian. Those cells which contain the plasmid will also have the resistance gene and survive the plating without a hitch. Those that do not will die, helping us filter out the unwanted cells. It’s a little experiment in microevolution every time! We also plate positive and negative controls, to make sure there were no errors made while making the plates. I imagine my plated bacteria to be like little pigs, all hogging out before we extract the bacon—the probe—a few days later.
If the cultures grow well, we are then able to transfer a colony to a liquid medium and grow up the selected stock. Creating liquid medium is an interesting process in itself, as all liquid and jars must be sterilized before use in bacterial transformations in a HUGE machine called an autoclave. The autoclave is like a gigantic high heat dishwasher and I’m terribly frightened of it. I managed to embarrass myself the last time I went in to collect my growing medium. My "procedure" for collecting medium after sterilization involves standing as far away from the freakish metal steam trap as I can while cranking the door open slightly, one foot outside of the autoclave room, then running into the hall as steam rushes out of the autoclave. Last time I spotted one of my coworkers giggling at the sight of me from across the hall. Ah well, better safe than sorry, with the autoclave. One must then wait ten minutes for all the steam to clear before collecting the sterilized equipment to avoid burns.
When the bacteria are done growing it's time to collect the "bacon." First we spin the liquid cultures in the mega centrifuge—a machine that looks rather like a washing machine (although I don't think my clothes would survive the 6,000 Gs). The pellet (solid at the bottom of the bottle) from the centrifugation is saved and the liquid growing medium is disposed. Next we lyse the bacterial cell walls with enzymes, and then expose the material to several washes and filters until all the bacterial DNA and cellular components are cleared and only the pure plasmid remains.
I still have yet to make probe from the plasmid, which is a story for yet another week.
And now, for those of you who are truly geeky, here’s a little section on how the ampicillin bacterial selection works.
~Antibiotic Mechanisms~
Like penicillin, ampilcillin functions by irreversibly inhibiting the enzyme transpeptidase, which is essential to the production of the peptidoglycan cell wall of E. coli bacteria.
Transpeptidase contains a serine amino acid at its active site, and it is this component that makes it susceptible to the ampicillin molecule.
In the picture below I’ve circled a four membered ring containing three carbons, a nitrogen, and an oxygen double bonded to a carbon.
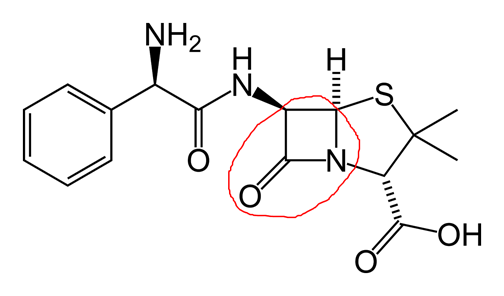
This ring is especially susceptible to nucleophilic attack as it is somewhat unstable. The alcohol of the serine molecule at the transpeptidase active site acts as a nucleophile, opening the ring. The hydrogen from the serine alcohol is transferred to the ring nitrogen and the carbonyl carbon binds to the alcohol oxygen of the serine molecule. The result is a disabled transpeptidase, covalently bound to the antibody:

This kills the bacteria, as they are unable to make and repair their cell walls without transpeptidase. That is, it kills all bacteria but those that have been transformed. The plasmid codes for the enzyme beta-lactamase, which acts on ampicillin in much the same way transpeptidase does. The serine alcohol of beta-lactamase attacks the ring, then interacts with water to produce an inactive antibiotic (the picture is for penicillin, but the mechanism against ampicillin is the same):
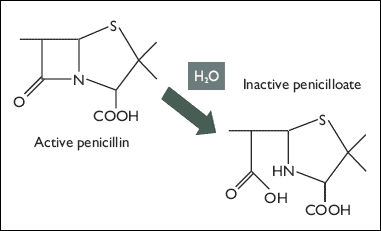
Sources:
Craig Stevens (lab mentor)
David Cole (biochemistry professor)
Ampicillin: wikipedia
Enzyme mechanism: http://www.antiinfectieux.org/antiinfectieux/PLS/Beta-lactames/PLS-Betalactames-mecanisme-action.html
Plasmid: http://teachline.ls.huji.ac.il/soreq/targil2/Q2-3.html
E. coli 1: http://www.rowett.ac.uk/edu_web/Gut_reactions3.html
E. coli 2: http://www.corante.com/loom/archives/evolution/

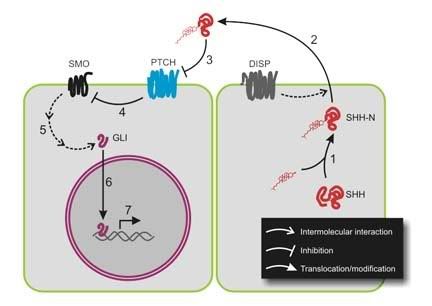
 Hedgehog proteins, in general, are best known for their role in regulating development by creating concentration gradients. The first
Hedgehog proteins, in general, are best known for their role in regulating development by creating concentration gradients. The first 

 Most of the brain does not hold the internal monologue that many of us perceive as consciousness. The constant "chatter" of thought over every detail would be too overwhelming. Rather, many of our thoughts go without saying; there is simply too much going on in the world to convey it all at once in the talkative regions of our cerebral cortex we call our conscious mind. (Imagination, too can be considered an internal monologue, just of the nonverbal variety)
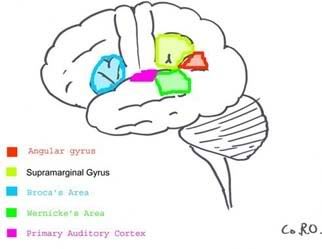
Most of the brain does not hold the internal monologue that many of us perceive as consciousness. The constant "chatter" of thought over every detail would be too overwhelming. Rather, many of our thoughts go without saying; there is simply too much going on in the world to convey it all at once in the talkative regions of our cerebral cortex we call our conscious mind. (Imagination, too can be considered an internal monologue, just of the nonverbal variety) One needs many parts of the brain functioning to see correctly, and blind sight often occurs in individuals who either have a portion their temporal lobe that deciphers what one is seeing destroyed or in individuals who have parts of their visual cortex damaged. According to a study by Stoerig and Cowey, even in monkeys where the entire visual cortex was experimentally removed blind sight was still found to occur. This means that seeing is not entirely a conscious act, rather only understanding what it is you’re looking at involves the conscious mind. Below is a diagram of the portions of the brain required for full conscious interpretation of a visual object:
One needs many parts of the brain functioning to see correctly, and blind sight often occurs in individuals who either have a portion their temporal lobe that deciphers what one is seeing destroyed or in individuals who have parts of their visual cortex damaged. According to a study by Stoerig and Cowey, even in monkeys where the entire visual cortex was experimentally removed blind sight was still found to occur. This means that seeing is not entirely a conscious act, rather only understanding what it is you’re looking at involves the conscious mind. Below is a diagram of the portions of the brain required for full conscious interpretation of a visual object: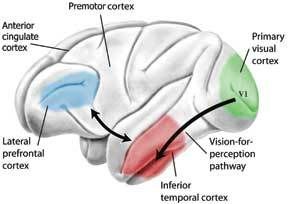
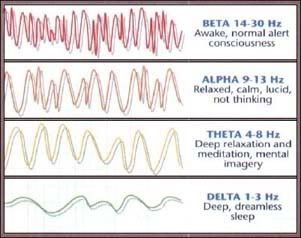 So how can we maximize the potential of our subconscious and bring it to surface in our conscious mind? According to studies using EEGs to record brain wave activity, the prime time for surfacing of these subconscious thoughts is during trance, meditation, right before sleep, or right after. These are times when the brain produces alpha waves; a characteristic pattern made when the subconscious, unconscious, and conscious patterns are all high and seem to be communicating best with each other.
So how can we maximize the potential of our subconscious and bring it to surface in our conscious mind? According to studies using EEGs to record brain wave activity, the prime time for surfacing of these subconscious thoughts is during trance, meditation, right before sleep, or right after. These are times when the brain produces alpha waves; a characteristic pattern made when the subconscious, unconscious, and conscious patterns are all high and seem to be communicating best with each other.