My Research on the Sonic Hedgehog Gene Mutation in Zebrafish!

Yesterday I had the wonderful opportunity to share my research with fellow scientists at the INBRE conference in Coeur d'Alene. My poster can be seen above (sorry you can't make out much more than the title) and I'd like to share with you all the juicy details about hedgehog genes and zebrafish today!
My entire summer thus far has been spent researching hedgehog (Hh) genes in zebrafish (Danio Rerio). Despite the name, hedgehog genes have little to do with hedgehogs, other than that they, like all vertebrates, possess them too. Hedgehog genes produce hedgehog proteins that play an important role in regulating the development of many cell types. They were so named for the mutation that led to their discovery; a mutation that would create a spiky, hedgehog appearance in fruit flies. In vertebrates, hedgehog proteins are known to play an important role in signaling from the midline of the developing organism, allowing it to maintain body symmetry as it grows, as apposed to a cycloptic look. Some studies have been done on their role in limb and trunk development, but my interest this summer has been the eye.
At some point in their evolutionary history, zebrafish had their genome duplicated and now have two versions of the hedgehog gene; one called sonic hedgehog and the other tiggy-winkle hedgehog (proof that smart-assed scientists get published too). This fact has some pretty sweet implications to research. Scientists have produced a sonic hedgehog knock-out mutant (called syu), which completely lacks the sonic hedgehog gene but retains tiggy-winkle. A complete absence of all hedgehog protein signaling would kill the embryo too soon, but allowing one hedgehog gene to remain while the other is absent will give us an opportunity to examine the effects of reduced hedgehog protein signaling on the development of the retina.
In previous studies, the rate of retinal mitosis was found to be reduced in syu mutants. In about half of the mutants, no cell differentiation occurred in the retina. In the half that did undergo differentiation, the spread of red and rod opsin (the receptor that reacts to various wavelengths of light) expression was very limited.
I had an opportunity this summer to look into the expression of other photoreceptor specific genes in syu mutants, including blue opsin and the transducins (the g-protein coupled to the opsin).
To study these mutants, fish must first be bred and sorted, which I have made a nifty diagram of below:
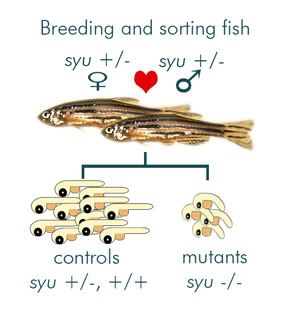
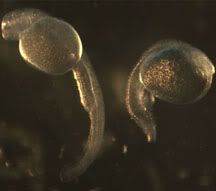
Since full mutants don’t live past 75 hours post fertilization (hpf), two zebrafish heterozygous for the mutation must be bred and the mutants are sorted out later by morphological differences. You can really see the difference below, at 50 hpf right after I dechorionate them (decorionate = pop open the eggs).
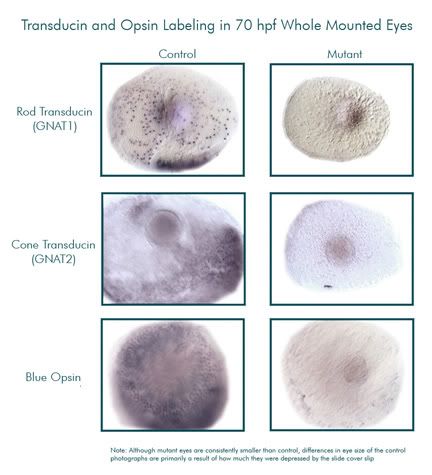
The embryos are fixed (killed and preserved) at 70 hpf, because this is an optimal time to look at photoreceptor development. Next, I used in situ hybridization to detect gene expression. In situ hybridization involves attaching complimentary RNA probe to messenger RNA (the first product of gene expression). We make the probes using E. coli bacteria, then extract the probe and introduce it into the tissue of the fixed embryo. Once exposed to the probe, the tissues have been prepared in such a way that will allow them to take up the probe and hybridize it to their mRNA. The next step involves attaching an antibody to that, then a dye is added that reacts with the antibody and produces purple coloration wherever gene expression is taking place. Below is a series of photos of the magnified eyes of mutant and control zebrafish. All the purple splotches and dots are where gene expression of our target genes is occurring. As you can see, the mutants had extremely reduced expression of these genes.
For each probe, I've tested 1 to 2 clutches of embryos. I hope to collect more embryos and produce additional data for statistical purposes. After that, I hope to look at how we can recover these lost photoreceptor cells by adding certain chemicals like retinoic acid or exogenous hedgehog proteins. This is exciting research because diseases that affect photoreceptor cells, such as macular degeneration, are a leading cause of blindness in the elderly population. Understanding how photoreceptors form by studying zebrafish may provide insight for future medical applications.

Yesterday I had the wonderful opportunity to share my research with fellow scientists at the INBRE conference in Coeur d'Alene. My poster can be seen above (sorry you can't make out much more than the title) and I'd like to share with you all the juicy details about hedgehog genes and zebrafish today!
My entire summer thus far has been spent researching hedgehog (Hh) genes in zebrafish (Danio Rerio). Despite the name, hedgehog genes have little to do with hedgehogs, other than that they, like all vertebrates, possess them too. Hedgehog genes produce hedgehog proteins that play an important role in regulating the development of many cell types. They were so named for the mutation that led to their discovery; a mutation that would create a spiky, hedgehog appearance in fruit flies. In vertebrates, hedgehog proteins are known to play an important role in signaling from the midline of the developing organism, allowing it to maintain body symmetry as it grows, as apposed to a cycloptic look. Some studies have been done on their role in limb and trunk development, but my interest this summer has been the eye.
At some point in their evolutionary history, zebrafish had their genome duplicated and now have two versions of the hedgehog gene; one called sonic hedgehog and the other tiggy-winkle hedgehog (proof that smart-assed scientists get published too). This fact has some pretty sweet implications to research. Scientists have produced a sonic hedgehog knock-out mutant (called syu), which completely lacks the sonic hedgehog gene but retains tiggy-winkle. A complete absence of all hedgehog protein signaling would kill the embryo too soon, but allowing one hedgehog gene to remain while the other is absent will give us an opportunity to examine the effects of reduced hedgehog protein signaling on the development of the retina.
In previous studies, the rate of retinal mitosis was found to be reduced in syu mutants. In about half of the mutants, no cell differentiation occurred in the retina. In the half that did undergo differentiation, the spread of red and rod opsin (the receptor that reacts to various wavelengths of light) expression was very limited.
I had an opportunity this summer to look into the expression of other photoreceptor specific genes in syu mutants, including blue opsin and the transducins (the g-protein coupled to the opsin).
To study these mutants, fish must first be bred and sorted, which I have made a nifty diagram of below:

Since full mutants don’t live past 75 hours post fertilization (hpf), two zebrafish heterozygous for the mutation must be bred and the mutants are sorted out later by morphological differences. You can really see the difference below, at 50 hpf right after I dechorionate them (decorionate = pop open the eggs).

The embryos are fixed (killed and preserved) at 70 hpf, because this is an optimal time to look at photoreceptor development. Next, I used in situ hybridization to detect gene expression. In situ hybridization involves attaching complimentary RNA probe to messenger RNA (the first product of gene expression). We make the probes using E. coli bacteria, then extract the probe and introduce it into the tissue of the fixed embryo. Once exposed to the probe, the tissues have been prepared in such a way that will allow them to take up the probe and hybridize it to their mRNA. The next step involves attaching an antibody to that, then a dye is added that reacts with the antibody and produces purple coloration wherever gene expression is taking place. Below is a series of photos of the magnified eyes of mutant and control zebrafish. All the purple splotches and dots are where gene expression of our target genes is occurring. As you can see, the mutants had extremely reduced expression of these genes.

For each probe, I've tested 1 to 2 clutches of embryos. I hope to collect more embryos and produce additional data for statistical purposes. After that, I hope to look at how we can recover these lost photoreceptor cells by adding certain chemicals like retinoic acid or exogenous hedgehog proteins. This is exciting research because diseases that affect photoreceptor cells, such as macular degeneration, are a leading cause of blindness in the elderly population. Understanding how photoreceptors form by studying zebrafish may provide insight for future medical applications.


3 Comments:
Looks like INBRE was fun, I missed it this year. Thanks for adding my blog. I will add yours as soon as I learn how to! hehe
By T, at 4:34 PM
T, at 4:34 PM
So freakin' exciting. Because of this research, maybe I won't be blind someday after all!
By Anonymous, at 10:41 PM
Anonymous, at 10:41 PM
Hi, I hope you don't mind, but I enjoyed your post so much, that I thought it would be a good addition to this month's Mendel's Garden carnival.
Check it out at: Discovering Biology in Digital World
By Sandra Porter, at 12:23 PM
Sandra Porter, at 12:23 PM
Post a Comment
<< Home